1. Introduction
The probability of gaining regulatory approval for newmedical therapies has decreased in latest years. On common, a brand new drugentering section I medical testing is estimated to have an 8% likelihood of reachingthe market, a lower from the historic fee of 14% . Major causes ofclinical trial failures embrace inadequate drug exercise (30%) andunacceptable toxicity profiles (30%) . The improvement of sturdy pharmacodynamic (PD)markers is essential for bettering the success of medicine in medical trials andwill information choice of an optimum drug dose to stability efficacy and toxicity PD markers are sometimes proximal in a molecularpathway to the drug goal and are used to measure the impact of a drugregardless of therapeutic impact. Another necessary part that contributesto the success of new therapies is the event of diagnostic biomarkersthat might permit higher affected person stratification.
Biomarkers present extra info at earlier stagesof the medical improvement course of, thus serving to to prioritize drug discoveryresources and permitting for higher early selections on the destiny of a developmentprogram. The US Food and Drug Administration (FDA) lately revealed severalwhite papers that acknowledge the significance of biomarkers in drug developmentand medical trials. While the FDA emphasised the necessity forbiomarkers to display goal neutralization, it additionally expressed tremendousinterest in codeveloping diagnostic markers to focus on the proper patientpopulation, thereby bettering the drug success fee The FDA additionally hasencouraged the adoption and integration of genomic information in drug improvement andregulatory evaluation , initiating and spearheading the MicroArray QualityConsortium (MAQC) challenge to evaluate key elements contributing to thevariability and reproducibility of microarray information. The MAQC has proven that microarray platformsare appropriate instruments to provide dependable, high-quality information that may assist drugdevelopment and regulatory choice making.
Systemic lupus erythematosus (SLE) is an autoimmunedisease that’s characterised by extreme immune system faults and theproduction of autoantibodies that result in irritation and tissue harm. The present normal of care entails the use ofcorticosteroids and poisonous immunosuppressive brokers which are extensively acknowledgedto trigger unacceptable hostile occasions with long-term use . Thus, novel therapies are wanted that immediately addressdisease pathogenesis with much less toxicity. Type I interferons (IFNs) have beenimplicated in the event of SLE for at the very least 25 years , and elevated ranges of IFN-α aredetected in the serum of some SLE sufferers . Previous outcomes from microarray research thatinvestigated gene expression profiles in the peripheral blood of SLE patientshave strengthened the concept kind I IFNs are concerned in diseasepathogenesis . Furthermore, assays reminiscent of real-time polymerasechain response (RT-PCR) have demonstrated that overexpression of IFN-α/β-inducible genescorrelated with elevated illness severity and exercise in SLE sufferers.
We are at the moment exploring an anti-IFN-α monoclonal antibody (mAb) as remedy for SLE and have used entire genome arrayanalyses to establish putative PD and diagnostic biomarkers to help in thedevelopment of the medical trial. Free IFN-α protein in the serum of SLEpatients can be probably the most cheap selection for a PD marker for evaluating ananti-IFN-α remedy inSLE. However, our inside research aswell as others present that solely a small fraction of SLE sufferers have measurableIFN-α protein in the sera . IFN-α-inducible genes,alternatively, are immediately downstream of the drug goal, are robustlyoverexpressed in entire blood (WB) of the bulk of SLE sufferers, and can bequantitatively measured by both microarray or TaqMan quantitative real-timereverse-transcriptase PCR- (QRT-PCR-) based mostly assays.
In this research, we’ve used the Affymetrix humangenome plus U133v2.Zero array platform to look at the magnitude and prevalence ofoverexpression of IFN-α/β-inducible genes in WB of SLE sufferers. Based onthese outcomes, we chosen a core group of IFN-α/β-inducible genesand confirmed the microarray outcomes utilizing TaqMan QRT-PCR. Furthermore, we used ex vivostimulation of wholesome donor peripheral blood mononuclear cells (PBMCs) withSLE affected person serum and subsequent neutralization with anti-IFN-α mAb oranti-IFN-α receptor (IFNAR) mAb to guage the contribution of IFN-α to theinduction of kind I IFN-inducible genes in WB of SLE sufferers.
2. Materials and Methods
2.1. Patients and Healthy Donor Controls
Two panels of SLE sufferers had been used in the research. The preliminary research panel included 41 SLE sufferers. WB from these SLE patientswas procured from Asterand (Detroit, Mich, USA),Cureline (South San Francisco, Calif,USA), and SeraCare (WestBridgewater, Mass, USA). All SLE sufferers had a historical past of at the very least 4 of 11 optimistic American College ofRheumatology (ACR) classification standards for the prognosis of SLE andactive illness manifestations on the time of pattern assortment. Thirty-nine (95%)had been girls, (imply ± SD age of 40 ± 15 years). Thirty-two of 33 (97%) sufferers who weretested for the presence of anti-nuclear antibodies (ANA) got here out optimistic. Thirty-one of 41 (76%) SLE sufferers had been at the moment receiving oral prednisone indoses starting from 1 to 30 mg/day, with 2 SLE sufferers additionally receiving pulseintravenous steroids. More than half (24/41) of SLE sufferers had been receiving atleast 1 different potential disease-modifying remedy: hydroxychloroquine(n = 10), cyclophosphamide (n = 6), methotrexate (n = 4), azathioprine (n = 2),cyclosporine (n = 1), or mycophenolate mofetil (n = 1).
The potential research panelincluded an unbiased set of SLE affected person samples that was used to demonstratea related distribution of sufferers with an overexpression of IFN-α/β-inducible genes. All sufferers accessible from a section 1a medical trial (MI-CP126) ofpatients had been used for the aim. Thispanel included WB from 54 SLE sufferers from MI-CP126 investigating anti-IFN-αmAb remedy in mild-to-moderate SLE. Patients (age ≥18 years) who met at the very least 4 of the 11 ACRcriteria for SLE had been enrolled in the trial. Stable SLE background therapies with acetaminophen, nonsteroidalanti-inflammatory medicine, antimalarials, and prednisone ≤ 20 mg/day or equivalentwere allowed.Patients who had been receiving cyclophosphamide, azathioprine,methotrexate, mycophenolate mofetil, cyclosporine, >20 mg/day prednisone (orequivalent), immunoglobulins, blood merchandise, investigational medicine, orantiviral therapies had been excluded, in addition to sufferers with energetic or chronicinfection, latest vaccination with stay attenuated viruses, latest herpeszoster, historical past of extreme herpes an infection, energetic central nervous systemlupus, clinically vital cardiac, cerebrovascular, liver, or renaldisease, or historical past of most cancers.Most sufferers had been middle-aged Caucasianfemales with delicate to reasonably energetic SLE with cutaneous involvement. Thestudy was carried out in response to the Declaration of Helsinki, and the studyprotocol was accepted by the institutional evaluation board at every website. Allpatients gave written knowledgeable consent earlier than study-related procedures wereperformed.
The management group consisted of WB from 24 healthynormal donors (age from 23 to 56; feminine: male ratio is roughly 5:1)enrolled internally (MedImmune, LLC.). All the blood donors gave writteninformed consent for the blood to be taken and used in this research. The majorityof the donors had been Caucasians.gives demographicinformation for the three teams described above.
All WB from SLE sufferers and controls had been collectedin PAXgene RNA tubes (PreAnalytiX GmbH) in response to the producer’sinstructions.
2.2. Total RNA Extraction and MicroarrayProcessing
AffymetrixHuman Genome U133 Plus 2.0 GeneChip arrays had been used in this research. Total RNAwas extracted from WB samples collected in PAXgene RNA tubes utilizing the QiagenPAXgene Blood RNA package (Valencia, Calif, USA). RNA purity and focus had been decided spectrophotometrically (260/280> 1.9). The era, fragmentation, and hybridization of biotin-labeledamplified complementary RNA (cRNA) had been carried out as outlined in the AffymetrixGeneChip guide (Santa Clara, Calif,USA). The hybridizations had been carried out in a single day and the washing/staining of arraysand scanning had been carried out in line with the usual Affymetrix protocol. Data seize and preliminary array high quality evaluation had been carried out with theGeneChip Operating Software.
2.3. Ex Vivo Stimulation of WB from HealthyDonors with Type I IFN Family Members
Ex vivo stimulation of WB was carried out on bloodcollected from Three wholesome donors enrolled internally (MedImmune, LLC.). Bloodsamples (6 mL) had been uncovered for therapies of automobile (1× PBS), a panel ofIFN-α subtypes (IFN-α2a, -4b, -5, -6, -7, -8, -10, -14, -16, -17), and IFN-β atconcentration of 3 × EC50. All the cytokines had been bought from PBL Biomedical(Piscataway, NJ, USA). Following dosing, the blood was incubated at 37°C, 5% CO2 for 4hours and transferred to a PAXgene RNA tube and inverted Eight to 10 instances. ThePAXgene tubes had been incubated at room temperature for 2 hours and then frozenuntil processed.
2.4. Microarray Data Analysis
ArrayAssistLite software program (Stratagene, La Jolla, Calif, USA)was used to calculate probe-level summaries (GC-RMA) from the array cellintensity information (CEL). Significanceanalysis of microarrays (SAMs) withcontrol of the false discovery fee was used to pick differentially regulatedgenes in SLE versus wholesome controls utilizing R packages (R Development Core Team, University of Auckland, New Zealand). Transcripts with a fold change ≥ 2 and q worth < 0.05 had been thought-about to be differentially regulated. Principalcomponents analyses (PCAs) and hierarchicalclustering analyses had been carried out utilizing SpotFire and R packages.
2.5. Pathway Analysis—GeneGo
Pathway and community evaluation of gene expression datawas carried out with the MetaCore built-in software program suite from GeneGo, Inc. (St. Joseph, Mich, USA) utilizing the genes decided to be significantlyregulated. The significance of regulation, given a specific pathway ornetwork, is approximated utilizing a hypergeometric distribution in which the P worth represents the likelihood of a specific gene set mapping arising bychance, given the (1) quantity of genes in the set of all genes on pathway maps,(2) genes on a specific pathway map, and (3) genes in the experiment.
2.6. TaqMan Low Density Array
The TaqMan Low Density Array (TLDA; AppliedBiosystems, Foster City, Calif, USA)was used to find out the fold-change differential for a panel of 18 genesbetween WB of 27 SLE sufferers and pooled RNA from 24 wholesome controls. Genesprinted on the array included: 9 kind I IFN-α subtypes (1, 2, 5, 6, 7, 8, 14,17, 21), Three further kind I IFNs (IFN-β, –κ, –ω), IFN-γ, IFNαR1, IFNαR2,IFNγR1, IFNγR2, and TNF-α. Double-stranded cDNA for every affected person pattern waspreamplified utilizing the TaqMan PreAmp Master Mix package (Applied Biosystems). Standard procedures for loading the array had been adopted and the array was runon a 7900HT Fast Real-Time PCR System (Applied Biosystems). Data evaluation ofthe ensuing Ct values was carried out with SDSv2.2.2 software program (AppliedBiosystems).
2.7. FluidigmBioMark System
A combination of 44 TaqMan Gene Expression Assays,together with Four reference management genes (Applied Biosystems), was ready usingthe TaqMan PreAmp Master Mix Kit (Applied Biosystems). A complete of 70 samples(35 from the 41 SLE sufferers in the unique research and 35 from the 54 SLEpatients in the possible research) had been run in triplicate (utilizing Three differentBioMark Real-Time PCR Systems) in opposition to a set of 48 TaqMan Gene ExpressionAssays in BioMark 48.48 dynamic array chips (Fluidigm Corp., South San Francisco, Calif, USA). Dynamic arrays had been loaded utilizing a NanoFlex 4-IFC Controller (Fluidigm Corp.)and real-time reactions had been carried out utilizing a BioMark Real-Time PCR System(Fluidigm Corp.). Results had been analyzed utilizing BioMark Real-Time PCR Analysissoftware. Cts above 20 had been excluded from the calculation. Delta-delta Cts (ΔΔCt) had been calculated utilizing the imply of Four referencegenes (GAPDH, TFRC, β2M, and 18S) and a calibrator pattern.
2.8. Ex VivoStimulation of PBMCs from Healthy Donors with Sera from SLE Patients
SLE serumsamples had been chosen based mostly on ranges of kind I IFN exercise as decided by areporter gene assay as beforehand described with some modifications. Briefly, HEK293H cells had been stablytransfected with a luciferase assemble (Gaussia princeps) underneath thecontrol of the IFN-stimulated response factor (ISRE). Transfected cells had been incubated with 50%affected person sera, and luciferase exercise was detected in the tradition supernatants24 hours later. Samples producing a sign larger than 1.5 instances of thenegative management (regular human serum) had been thought-about optimistic. To decide whetherIFN-α wasresponsible for the optimistic response, cells had been handled with an anti-IFN-α mAb (humanIgG1; MedImmune, LLC.)and % neutralization was calculated. Serum samples had been chosen for exvivo stimulation of wholesome donor PBMC based mostly on their stage of IFN-α exercise.
PBMCs wereharvested from WB of a wholesome volunteer utilizing Ficoll-Pacque gradientcentrifugation in response to producer’s directions (GE Life Sciences,Uppsala, Sweden) and had been resuspended in RPMI 1640 media with GlutaMAXcontaining 10% fetal bovine serum (Invitrogen, Carlsbad, Calif, USA). Tomeasure the results of SLE serum on the wholesome donor cells, PBMCs werecultured at a density of 5 × 106 cells/mL in 250 μL/properly of a 24-wellplate containing 25% SLE affected person serum, in the presence or absence of thefollowing neutralizing antibodies: anti-human-IFN-α (0.1, 1, and 10 μg/mL; humanIgG1; MedImmune, LLC.),anti-human IFN-γ (10 μg/mL; mouse IgG1, clone MMHG-1; PBL), anti-human-IFNAR1 (10 μg/mL; human IgG1; MedImmune, LLC.), and anti-HIVgp120 as a unfavourable management (10 μg/mL; humanIgG1, MedImmune, LLC.). Following 4-hour incubation at 37°C,cells had been handled with Trizol LS (Invitrogen) and saved at −70°Cfor subsequent RNA isolation.
In a pilot research, we noticed that the identical SLE serumsample elicited very comparable responses in inducing the overexpression ofIFN-α/β-inducible genes in PBMCs from Three wholesome donors (information not proven). Thisallowed us to restrict the assay to 1 wholesome donor PBMCs in order that extra SLE serumsamples could possibly be included in the research. Therefore, we chosen 6 SLE serumsamples based mostly on their IFN-a bioassay outcomes described above and used thesesamples to stimulate PBMCs from one wholesome donor. This supplied a complete of 42 microarray experiments (i.e., 6 sera samplesfrom SLE sufferers × 7 situations).
3. Results
3.1. Ex Vivo Stimulation of Healthy Donor WB with IFN-α Subtypesand IFN-β
To decide theprevalence and magnitude of the overexpression of IFN-α/β-inducible genesin WB of SLE sufferers, we first carried out ex vivo stimulation of healthydonor WB with completely different members of the sort I IFN household. For every trio of cytokine therapies, a paired Student’s t-test and common fold change wascalculated between the three cytokine therapy replicates and the three untreatedhealthy donor WB samples. Only these probes that exhibitedat least a 2-fold change and P < .05throughout all cytokine therapies had been retained (the small sizes in eachcomparison restricted the use of a number of testing adjustment). We noticed that807 and 562 transcripts had been uniformly upregulated and downregulated,respectively, after stimulation of WB of Three wholesome donors with every of 10 IFN-αsubtypes or IFN-β for Four hours.
3.2. Overexpression of IFN-α/β-Inducible Genes is Robust and Prevalent in WB of SLEPatients
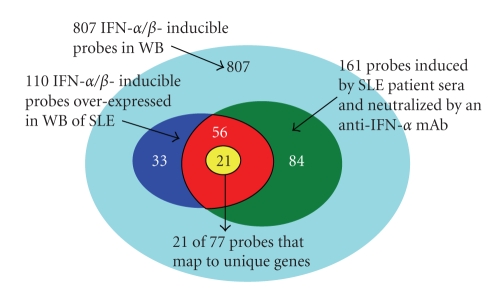
To identifycandidate PD markers for anti-IFN-α mAbclinical trials in SLE, we utilized the Affymetrix array platform to profile WBfrom 41 SLE sufferers in the preliminary research and 24 wholesome donors. We observedthat 239 and 88 transcripts had been upregulated and downregulated, respectively,in WB of SLE sufferers in contrast with wholesome controls. Of the 239 transcriptsupregulated in WB of SLE sufferers, 110 had been IFN-α/β-inducible (asdefined by ex vivo stimulation of WB with kind I IFN relations).
A totalof 30/41 of the SLE sufferers profiled confirmed vital overexpression of theIFN-α/β-inducible genesignature. To quantify the magnitude of overexpression of IFN-α/β-inducible genes in WB of SLE sufferers, we developed an algorithm that takes benefit of the wholegenome array strategy. Briefly, we chosen the 25 most extremely overexpressed IFN-α/β-inducible genes inindividual SLE sufferers based mostly on the 807 IFN-α/β-inducible transcripts generated from the ex vivo stimulation of wholesome donorWB research, and used the median fold change of these 25 genes to assemble anIFN-α/β-inducible genesignature rating for every SLE affected person. reveals thedistribution of the IFN-α/β-inducible genesignature scores of the 41 SLE sufferers in the preliminary research. We categorised theSLE sufferers into Three teams based mostly on their IFN-α/β-inducible genesignature rating: excessive IFN-α/β-inducible genesignature (rating > 10); reasonable IFN-α/β-inducible genesignature (rating 4–10); and weakIFN-α/β-inducible genesignature (rating < 4). The classification of SLE sufferers based mostly on IFN-α/β-inducible genesignature rating is principally for the aim of evaluating PD in the early phasesof medical trials of anti-IFN-α mAb remedy in SLE. The SLE sufferers with a weakor nondetectable IFN-α/β-inducible genesignature rating are unlikely to supply correct evaluation of thepharmacologic impact of anti-IFN-α mAb in these sufferers.